Scientists Map All Possible Drug Compounds in ‘Small Molecule Universe’
Researchers at Duke University have developed a digital library of all the small carbon-containing molecule structures that can possibly be developed as drugs— even ones that don't exist yet. The results provide "a near-infinite source of diverse novel compounds" that could allow chemists to identify promising structures for new drugs.
Using a new computerized mathematical model they called the Algorithm for Chemical Space Exploration with Stochastic Search (ACSESS), researchers led by chemist David Beratan mapped all the possible organic structures in the "small molecule universe," which is estimated to contain over 1 novemdecillion chemical compounds— that's 10^60, or 1 with 60 zeroes after it.
"The small-molecule universe is astronomical in size," Beratan told Duke Today, the university newsletter. "When we search it for new molecular solutions, we are lost. We don't know which way to look."
The molecular map created with ACSESS can help scientists navigate the vast field of possible compounds in order to see which chemical structures have already been developed, and how they might build the shapes in the unexplored "white space."
The researchers describe their algorithm and accompanying small molecule universe representative map (SMU-RUL) in a paper published this month in the Journal of the American Chemical Society.
Beratan told Duke Today that molecular solutions for many global issues can be found in the unexplored chemical space, "whether it's a cure for disease or a new material to capture sunlight." Developing all the compounds in that space would be prohibitively expensive and time-consuming, but the ACSESS model makes it easier to identify structures with useful properties.
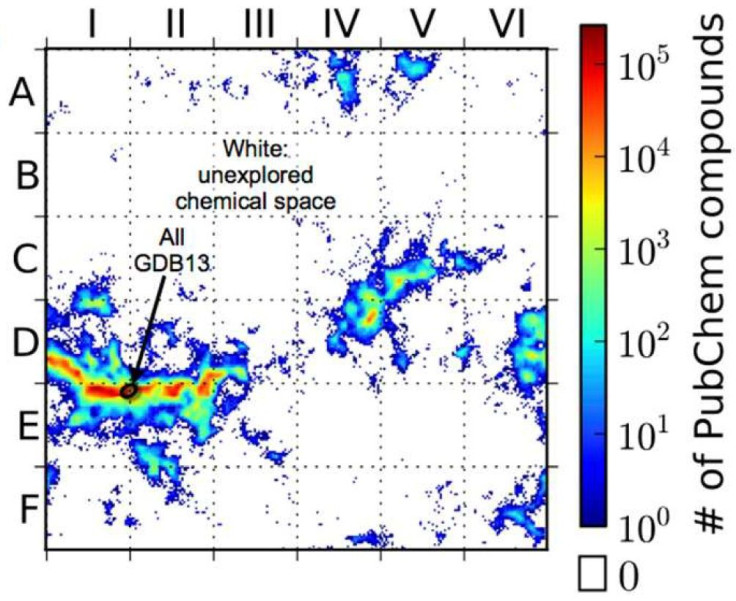
Digital molecular libraries have already yielded advances in chemistry research. The "GDB13" database, for example, which maps almost 1 billion possible organic compounds with 13 or fewer atoms, has led to several successful new drugs since it was released in 2009. Other methods expand on existing drug structures to model potential new ones.
The ACSESS library, however, dramatically expands the diversity of potential chemical compounds, and maps entirely new regions of the small molecule universe.
Postdoctoral associate Aaron Virshup programmed the algorithm to create a digital library of almost 9 million structures that represented different regions of the small molecule universe, then make random "chemical mutations" that can change the structure of each compound. The algorithm also accounts for whether a chemical structure is unstable, to limit the library to compounds that can realistically be created.
"The idea was to start with a simple molecule and make random changes, so you add a carbon, change a double bond to a single bond, add a nitrogen," Virshup explained to Duke Today. With enough repetitions of the process, researchers can use ACSESS to get to any possible molecule.
The Duke team then created a self-organizing map of the structures revealed by the ACSESS model, which they compared to all the chemical compounds that exist in the PubChem database. The resulting map revealed enormous swaths of unexplored chemical space, ripe for mining by scientists.
"With the map, we can tell chemists, if you can synthesize a new molecule in this region of space, you have made a new type of compound," Virshup told Duke Today. "If you're in the blank spaces on our small molecule map, you're guaranteed to make something that isn't patented yet."